Motivation
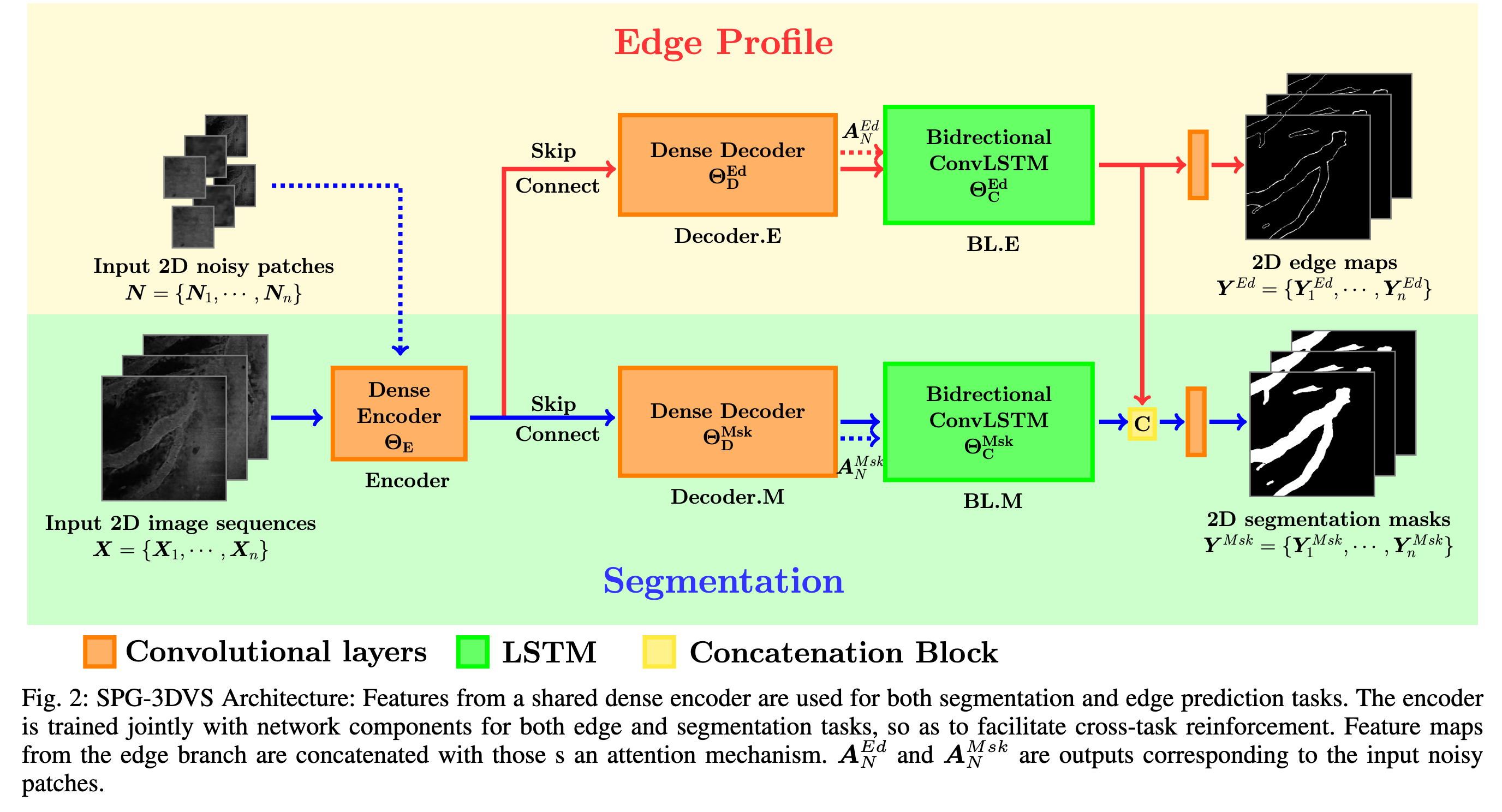
Network Structure
Loss Function
Classification Loss:
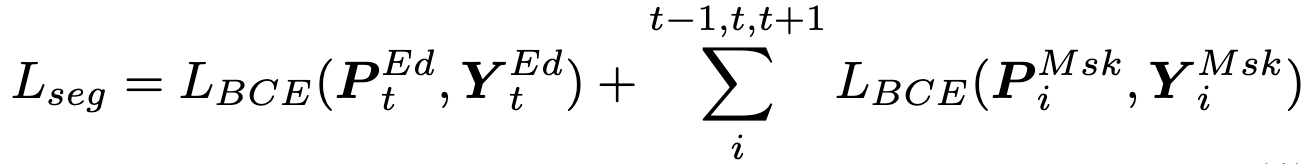
Domain Specific Regularization:
1. Data Adaptive Noise Robustness:
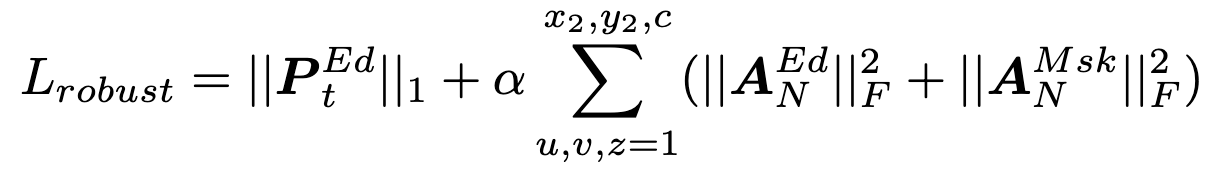
2. Local Homogeneity:
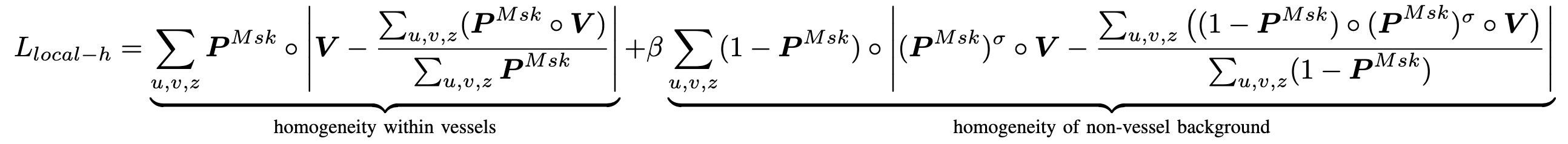
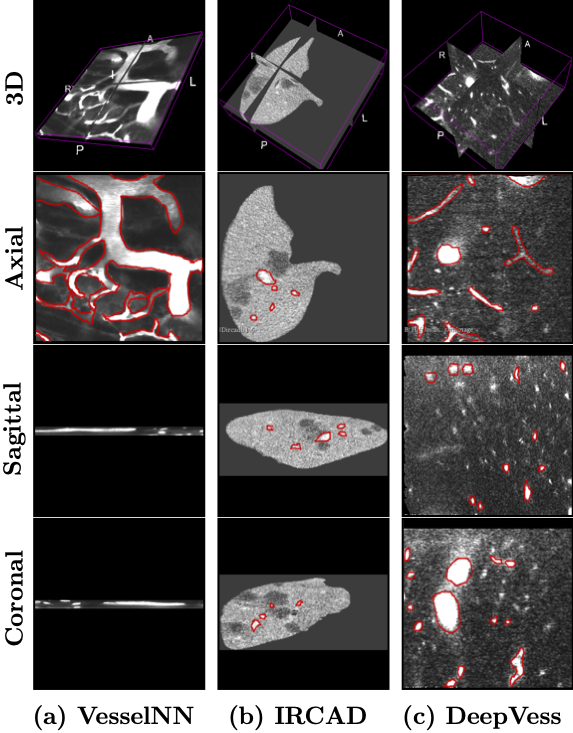
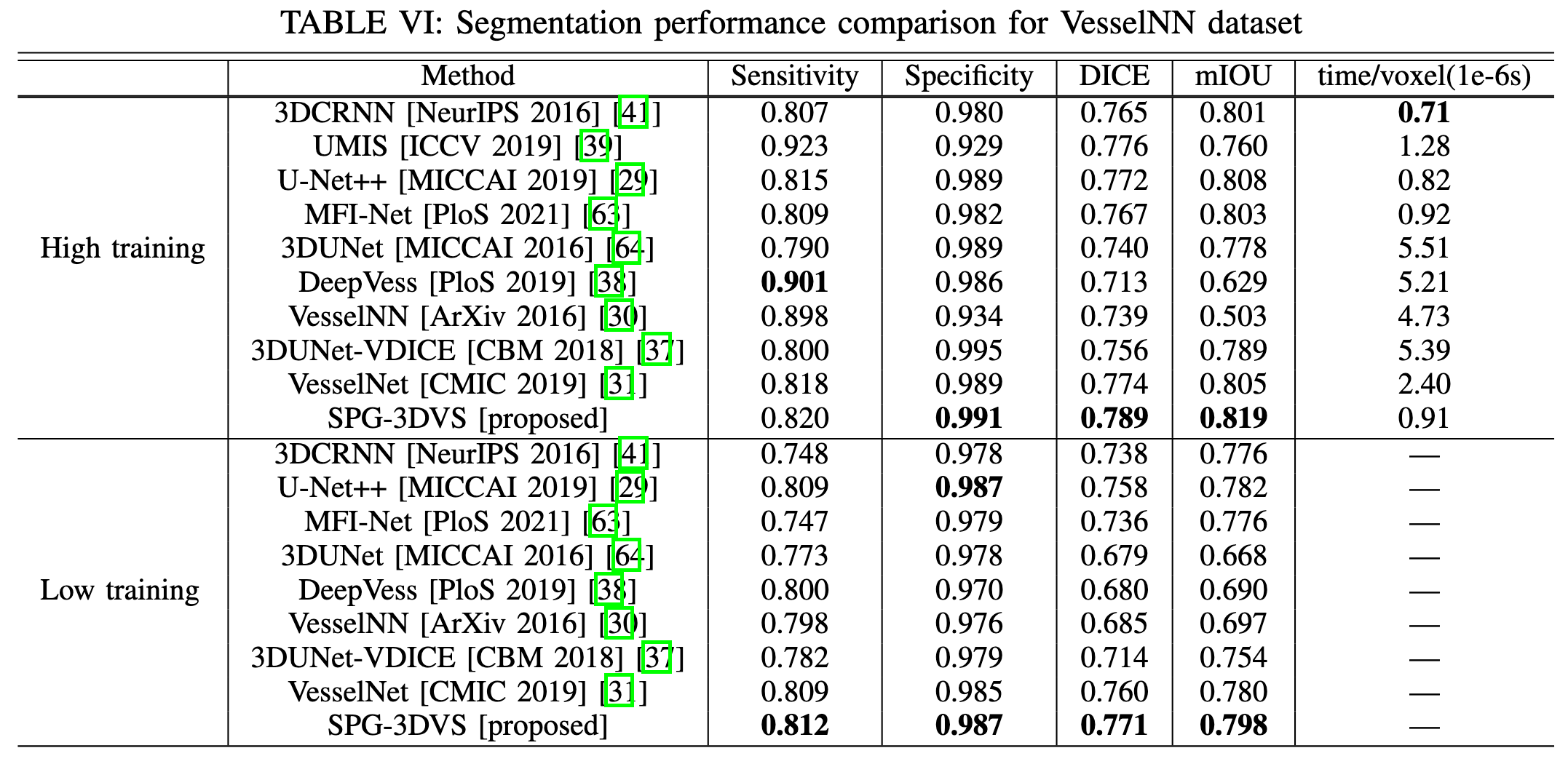
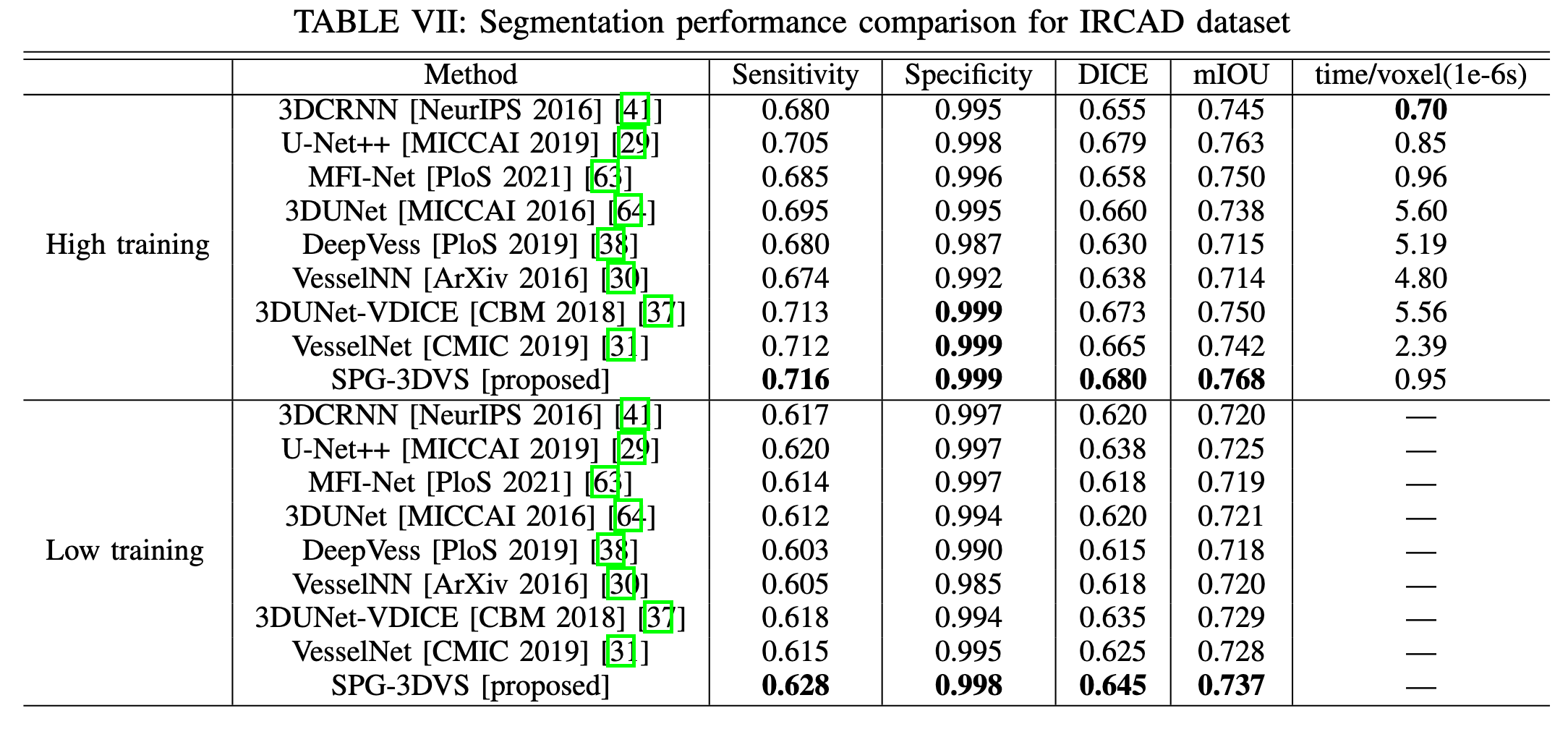
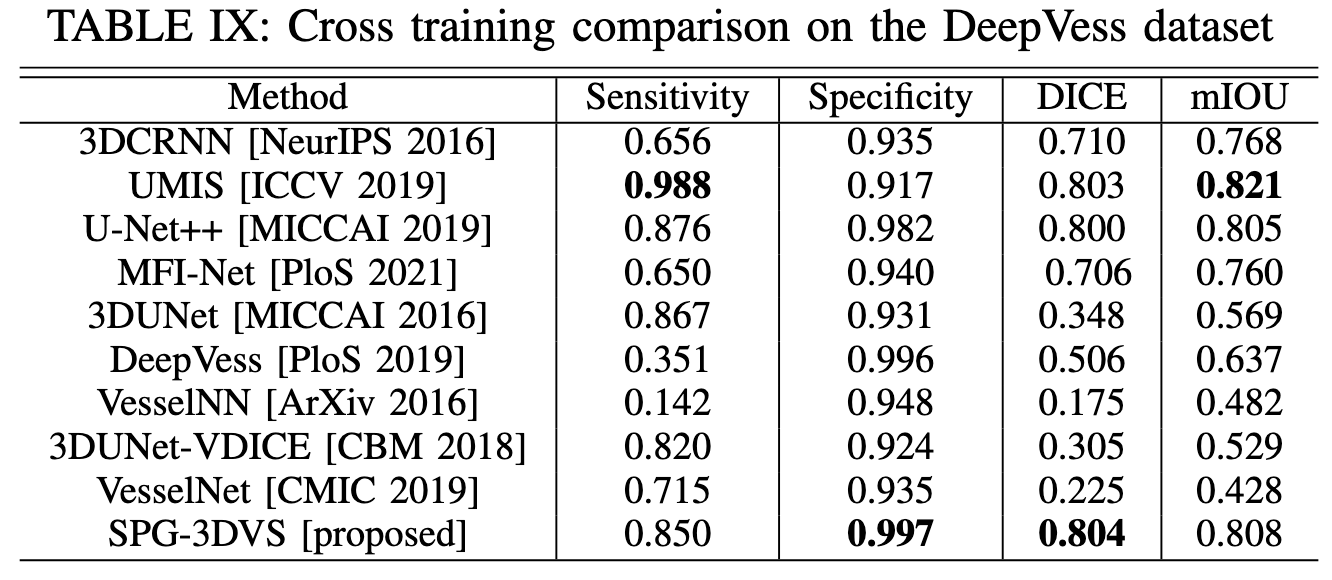
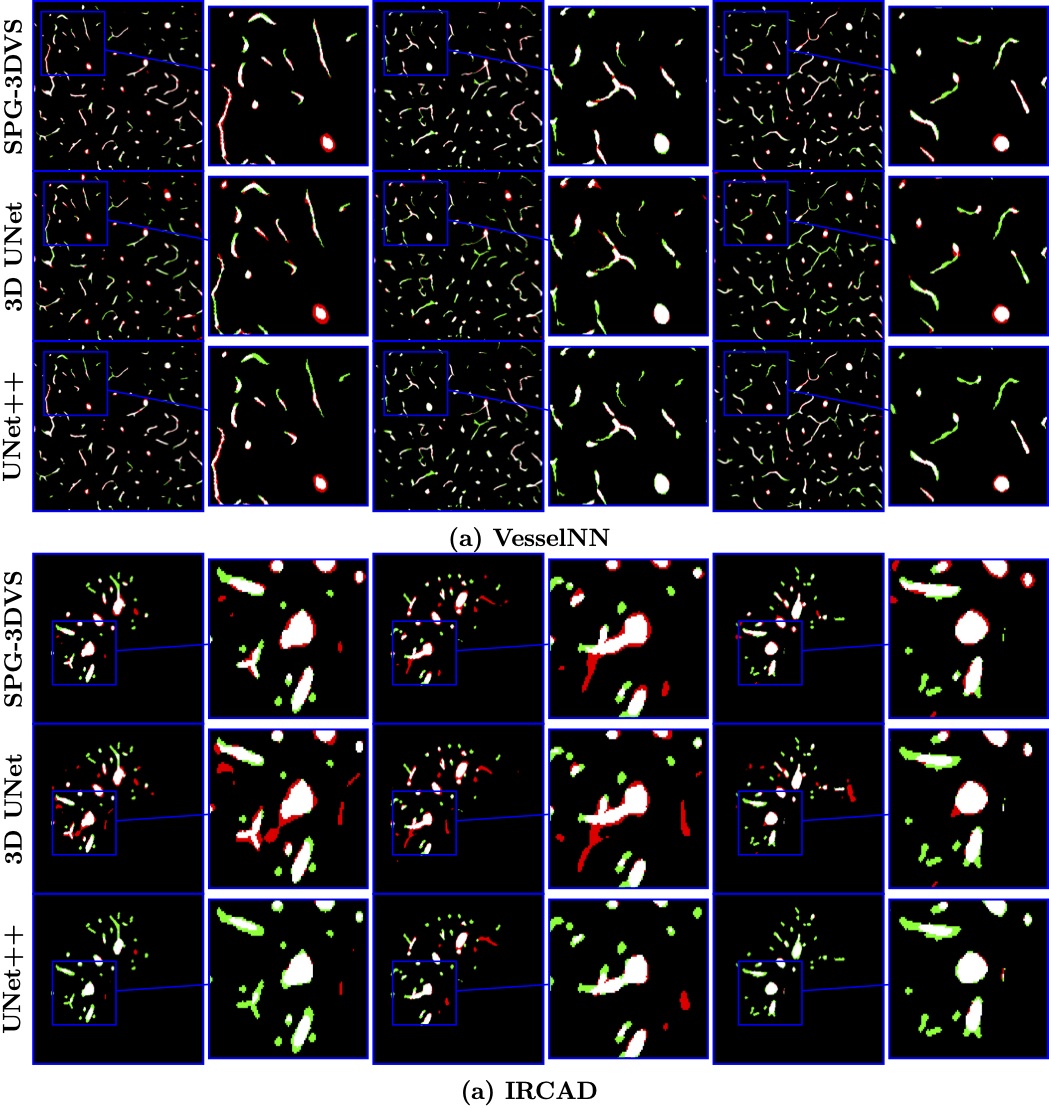
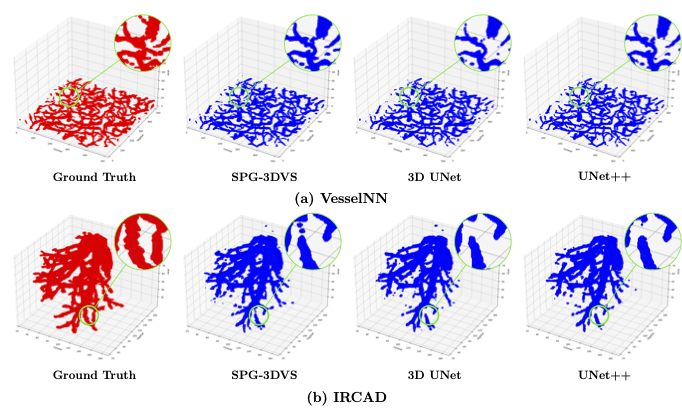
Experimental Results
Code
The code, you can find it in [this Github repository].
code, you can find it in [this Github repository].
Any feedback is welcome.
Related Publications
X Li, R Bala, V Monga, "Structural Prior Models for 3-D Deep Vessel Segmentation", in IEEE International Conference on Acoustics, Speech, and Signal Processing, 2022. [IEEE Xplore]
X Li, R Bala, V Monga, "Robust Deep 3-D Blood Vessel Segmentation Using Structural Priors", in IEEE Transactions on Image Processing, 2022. [IEEE Xplore]
Selected References
H Cui, X Liu, N Huang, "Pulmonary vessel segmentation based on orthogonal fused U-Net++ of chest CT images", in International Conference on Medical Image Computing and Computer-Assisted Intervention, 2019. [IEEE Xplore]
O. Çiçek, A. Abdulkadir, S. S. Lienkamp, T. Brox, and O. Ronneberger, "3D U-Net: Learning dense volumetric segmentation from sparse annotation", in International Conference on Medical Image Computing and Computer-Assisted Intervention, 2016. [IEEE Xplore]

