Histopathological
Image Data Sets
People
Please contact Prof.
Vishal Monga, Department of Electrical Engineering, Pennsylvania State
University, for information regarding this research.
Related papers
1.
U.
Srinivas, H. S. Mousavi, C. Jeon, V. Monga, A. Hattel,
and B. Jayarao, “SHIRC: A simultaneous Sparsity model
for Histopathological Image Representation and Classification,” in Proc. IEEE
International Symposium on Biomedical Imaging, San Francisco, April 2013.
2. U. Srinivas, H. S. Mousavi, V. Monga, A. Hattel, and B. Jayarao,
“Simultaneous sparsity model for histopathological image representation and
classification,” IEEE Transactions on Medical Imaging, under review.
Software
The MATLAB code
corresponding to our proposed simultaneous sparsity model can be downloaded here.
Data Sets
Two different histopathological image data sets are discussed here:
1.
ADL data set
2.
IBL data set
ADL Data Set
The images in this data set have been acquired by pathologists at the
Animal Diagnostics Lab (ADL), Pennsylvania State University.
Representative images from the data set can be downloaded here. Features of the data set:
1.
Hematoxylin-eosin
(H&E)-stained tissues
2.
Scanning
at 40x optical magnification using a whole slide digital scanner
3.
Three
bovine organs:
·
Kidney
(healthy/inflamed)
·
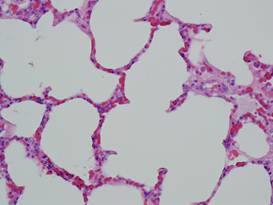
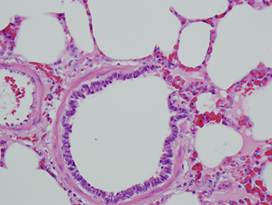
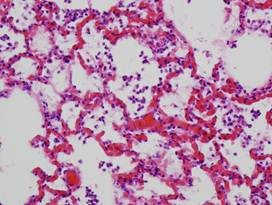
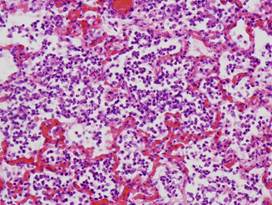
Lung
(healthy/inflamed)
·
Spleen
(healthy/inflamed)
4.
120
images per condition per organ
5.
Ground
truth labels for healthy and inflammatory tissue obtained via manual detection
and segmentation
|
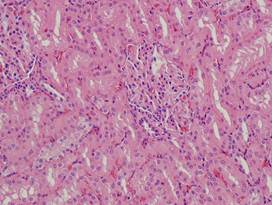
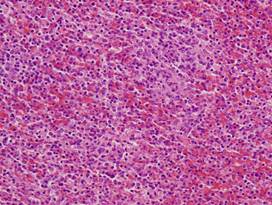
(a)
Healthy
lung |
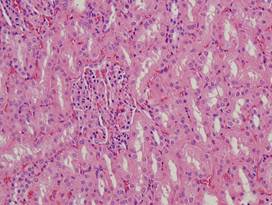
(b)
Healthy
lung |
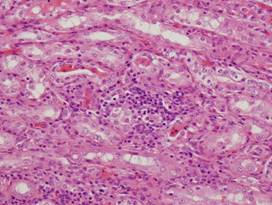
(c)
Inflamed
lung |
(d)
Inflamed
lung |
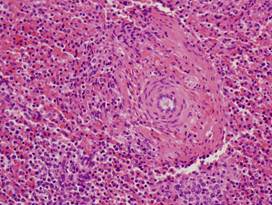
|
(e)
Healthy
kidney |
(f)
Healthy
kidney |
(g)
Inflamed
kidney |
(h)
Inflamed
kidney |
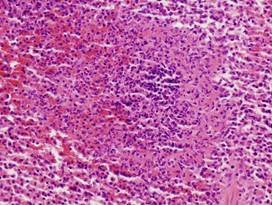
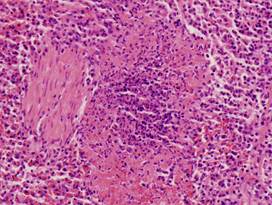
|
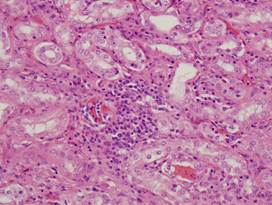
(i)
Healthy
spleen |
(j)
Healthy
spleen |
(k)
Inflamed
spleen |
(l)
Inflamed
spleen |
IBL Data Set
This data set contains images of human intraductal
breast lesions (IBL). They have been acquired by Clarion Pathology Lab,
Indianapolis and the Computer and Information Science Department, Indiana
University-Purdue University Indianapolis (IUPUI), Indiana. Features of the
data set:
1.
Two
well-defined categories: usual ductal hyperplasia (UDH) and ductal carcinoma in
situ (DCIS)
2.
Ground
truth labels assigned manually by pathologists
3.
Total
of 120 regions of interest (RoIs), or equivalently
images, used for experiment: 60 training, 60 test
Images cannot be made publicly available. Please contact Prof. Murat Dundar
at IUPUI for additional information.